Abstract
Background
Chronic lymphocytic leukemia (CLL) involving tissues is characterized by sheets of small lymphocytes and vague nodules of larger cells, known as proliferation centers (PCs). CLL can undergo progression to accelerated CLL (aCLL) or further progress to diffuse large B-cell lymphoma, also known as Richter transformation (RT). Distinguishing CLL with many PCs from aCLL or RT can be challenging, particularly in small needle-biopsy specimens. In this study, we sought to design an artificial intelligence (AI)-based tool to automate and enhance the delineation of PCs and provide an objective approach to CLL/SLL acceleration/transformation.
Material & Methods
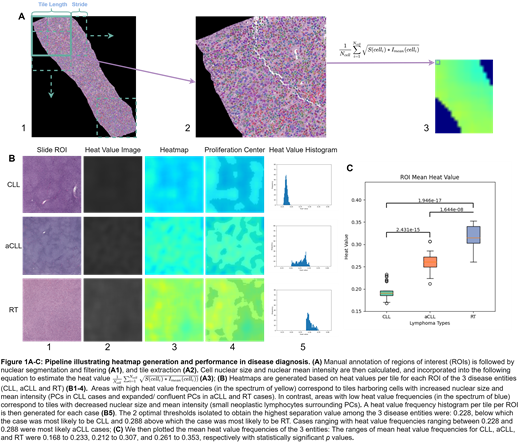
We manually annotated 25, 28 and 21 regions of interest (ROIs) encompassing small round PCs and confluent/ expanded PCs of 10 CLL, 12 aCLL, and 8 RT digitized hematoxylin & eosin stained slides, respectively (Figure A1). We analyzed the ROIs with both length and width larger than 2,000 pixels and set the tile length and stride as 1,000 and 100 pixels, respectively (Figure A1), and were able to extract sufficient tiles from each ROI (Figure A2). To recreate PCs, after performing nuclear segmentation via convolutional neural network and quality control, we quantified the nuclear size/intensity of cells occupying each tile (Figure A3). Nuclear size varied from 8 to 108 square micrometers, and nuclear mean intensity varied from 0 to 255. We normalized both the values of nuclear size and mean intensity to 0.0 and 1.0, by subtracting the minimum value and dividing it by the value range length. Nuclear size and mean intensity were represented as S(celli) and Imean(celli), respectively. We estimated the heat value of one tile integrating nuclear and mean intensity using the equationin Figure A2.
Results
We generated heatmaps based on the heat values per tile inside each ROI from the 3 disease entities (CLL, aCLL and RT), as illustrated in Figures B1-4. Areas with high heat values (in the yellow spectrum) correspond to tiles harboring cells with increased nuclear size and mean intensity (PCs in CLL cases and expanded/ confluent PCs in aCLL and RT cases). In contrast, areas with low heat values (in the blue spectrum) correspond to tiles with decreased nuclear size and mean intensity (small neoplastic lymphocytes surrounding PCs) (Figure B4).We then generated a heat value histogram per tile for each ROI (Figure B5).Based on these results, the two optimal thresholds isolated to obtain the highest separation value among the three disease entities based on the optimal F-score were: 0.228, below which the case was most likely to be CLL, and 0.288, above which the case was most likely to be RT. Cases with heat values ranging between 0.228 and 0.288 were most likely aCLL cases. We then plotted the mean heat value frequencies of the 3 entities: There was a significant difference in the ranges of mean heat value frequencies for CLL, aCLL, and RT, which were 0.168 to 0.233, 0.212 to 0.307, and 0.261 to 0.353, respectively (Figure C). The accuracy and area under the curve diagnostic predictive values using data from nuclear size alone were 0.658 (+/-0.115) and 0.771 (+/-0.096), respectively; and using mean nuclear intensity, 0.679 (+/-0.094) and 0.841 (+/-0.052), respectively; with a noticeable increase using heat value frequencies (integrating the nuclear size and mean nuclear intensity) reaching 0.813 (+/-0.0630) and 0.885 (+/-0.109), respectively.
Conclusion
We describe a novel AI-based heatmap technique to objectively assess the extent of PCs in CLL, based on the integrative analysis of cell nuclear size and mean nuclear intensity. We suggest that an ROI mean heat value less than 0.228 is predictive of CLL, and more than 0.288 is predictive of RT. aCLL cases demonstrate a mean heat value ranging from 0.228 to 0.288. Using the mean heat value of all cases, we were able to reliably separate the three entities in question with robust diagnostic predictive values.
Khoury: Stemline Therapeutics: Research Funding; Kiromic: Research Funding; Angle: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal